CryoSPARC v4.4.1 Now Available: Featuring one-click workflows, reference based motion correction, orientation diagnostics and much more!CryoSPARC v4.4.1 Now Available: Featuring one-click workflows, reference based motion correction, orientation diagnostics and much more! Learn more
Coronavirus spike protein structure solved using cryoSPARC
February 19, 2020Researchers from the University of Texas at Austin and the National Institutes of Health recently published a study in Science detailing the first ever cryo-EM structure of the 2019-nCoV spike (S) glycoprotein in the prefusion conformation.
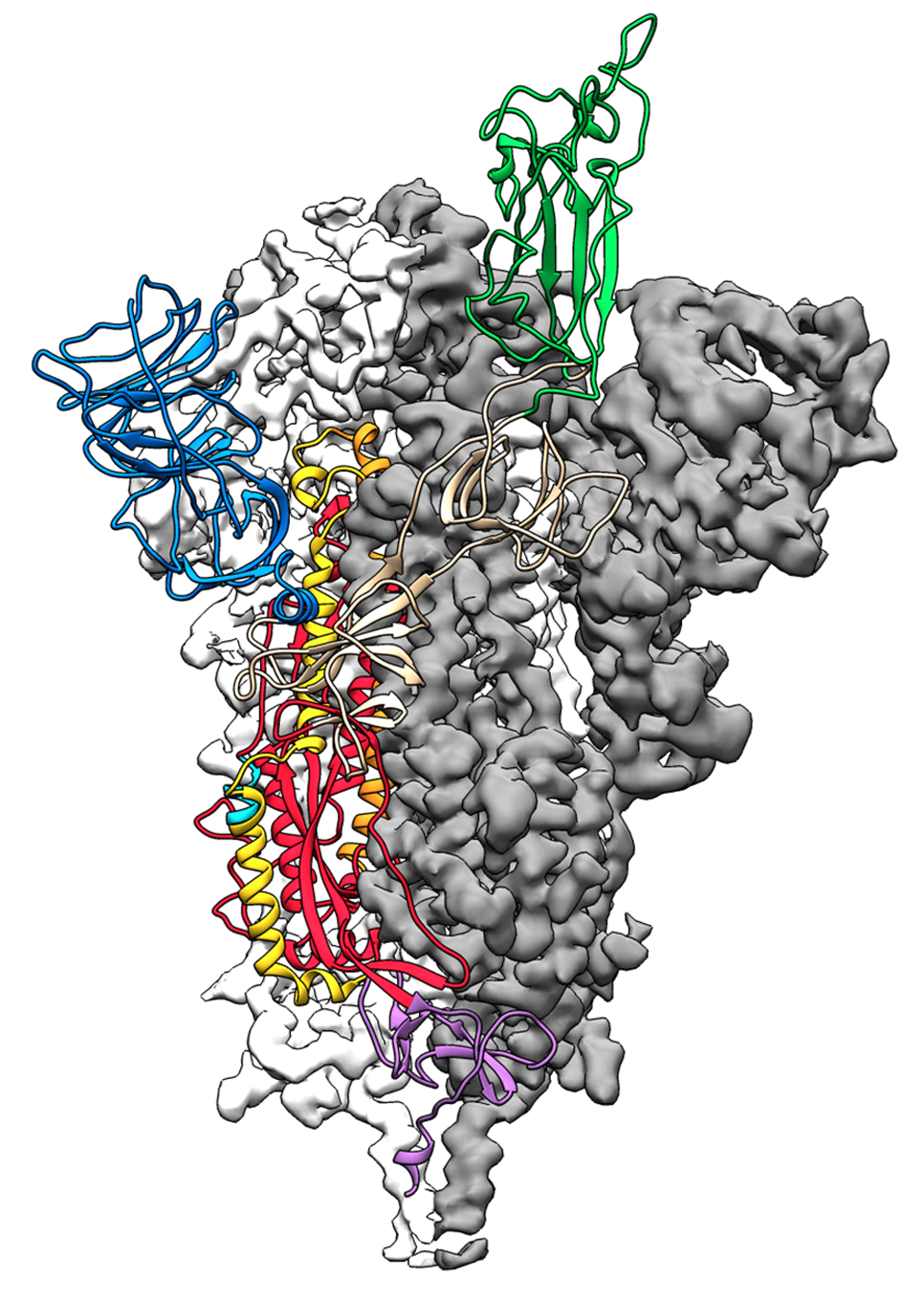
Cryo-EM structure of the 2019-nCoV spike protein. Image source
The novel coronavirus 2019-nCoV causes fever, severe respiratory illness and pneumonia and is part of an outbreak affecting thousands of individuals globally. Obtaining structural information on the virus and especially on the spike protein, which is the part that attaches to and infects human cells, is a critical breakthrough on the road to vaccine development.
The UT Austin/NIH team led by Dr. Jason McLellan, received the genome sequence of the virus from another team. They performed cryo-EM analysis and reconstructed the structure, all within 12 days. Their manuscript was expedited for review by Science and published online just a few days after they submitted a pre-print on bioRxiv.
The team performed 2D classification, 3D classification, non-uniform refinement and 3D variability analysis in cryoSPARC v2.
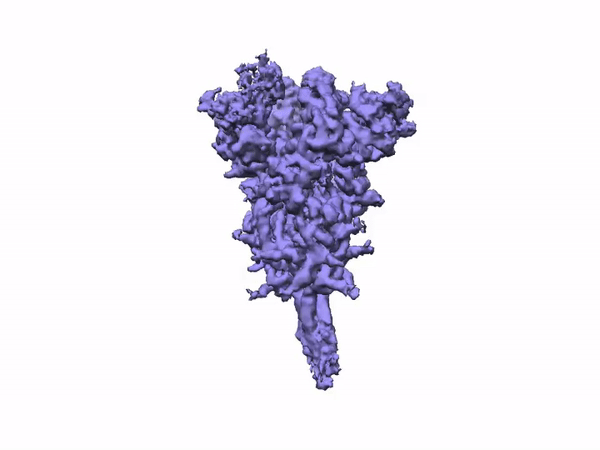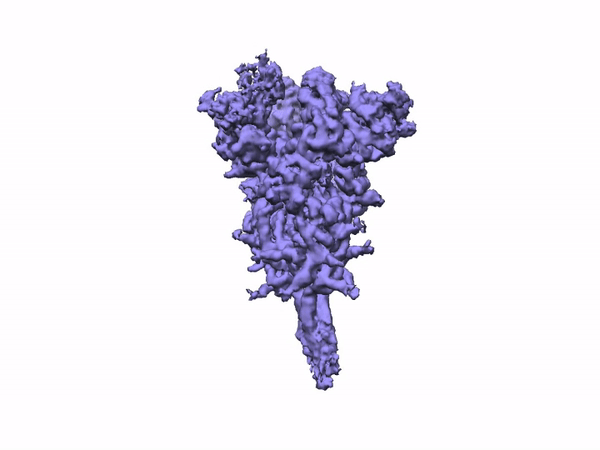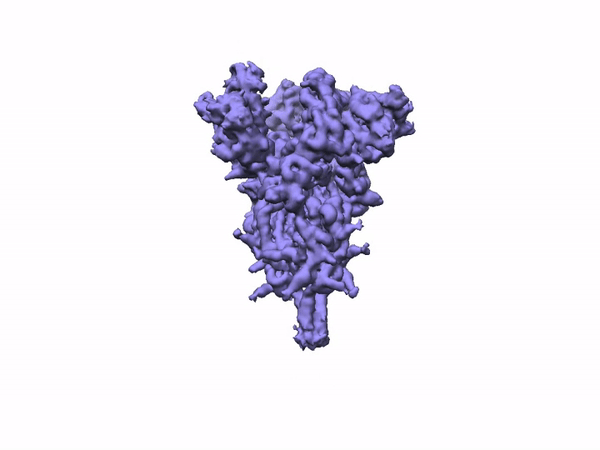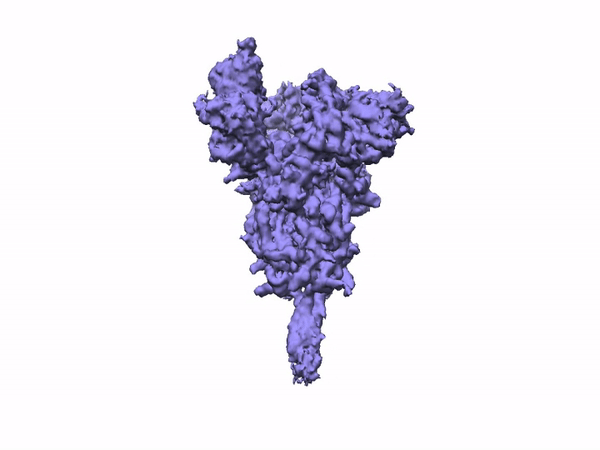
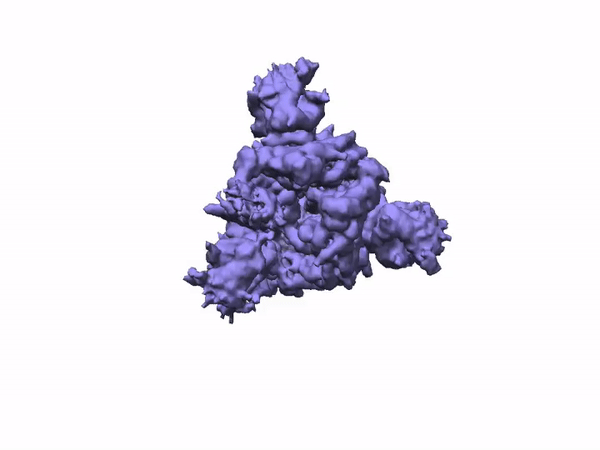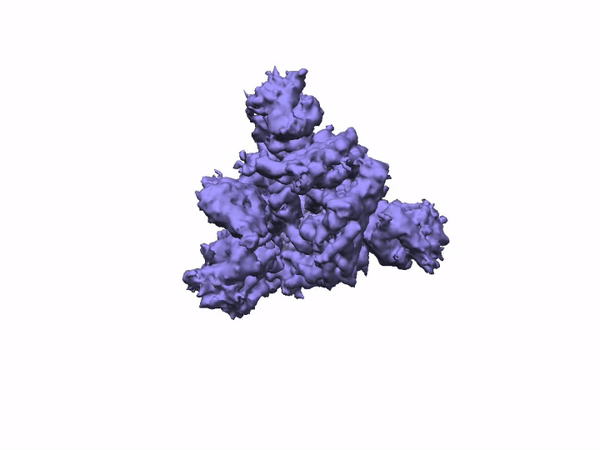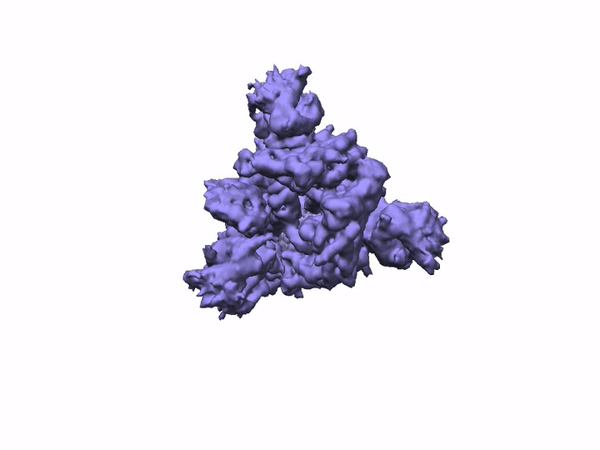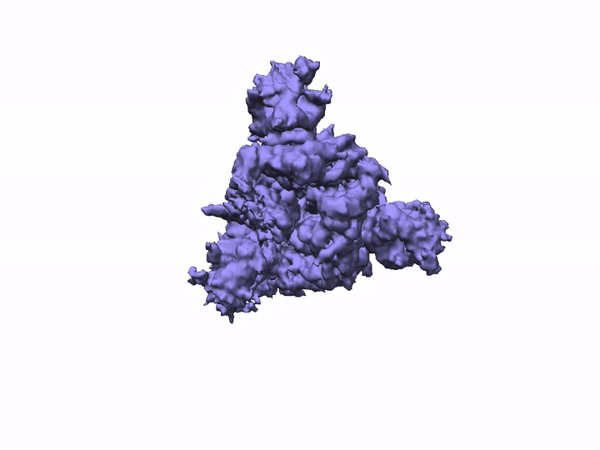
3D Variability Analysis in cryoSPARC v2 resolves flexible conformational change of the S-protein. Video source
Useful links:
- Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science eabb2507, https://doi.org/10.1126/science.abb2507 (2020).
- Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV Spike in the Prefusion Conformation. biorXiv, doi: https://doi.org/10.1101/2020.02.11.944462