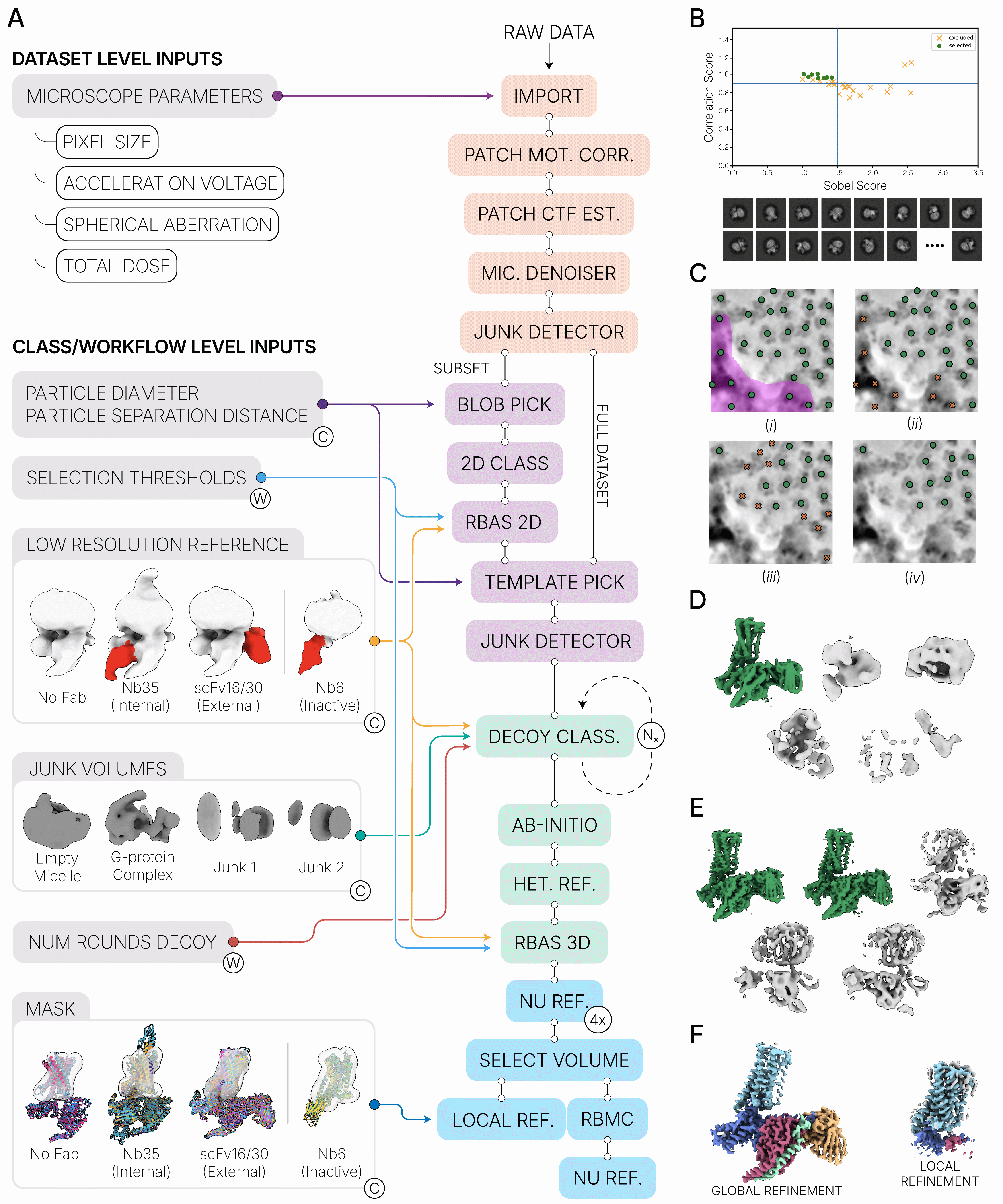
Overview
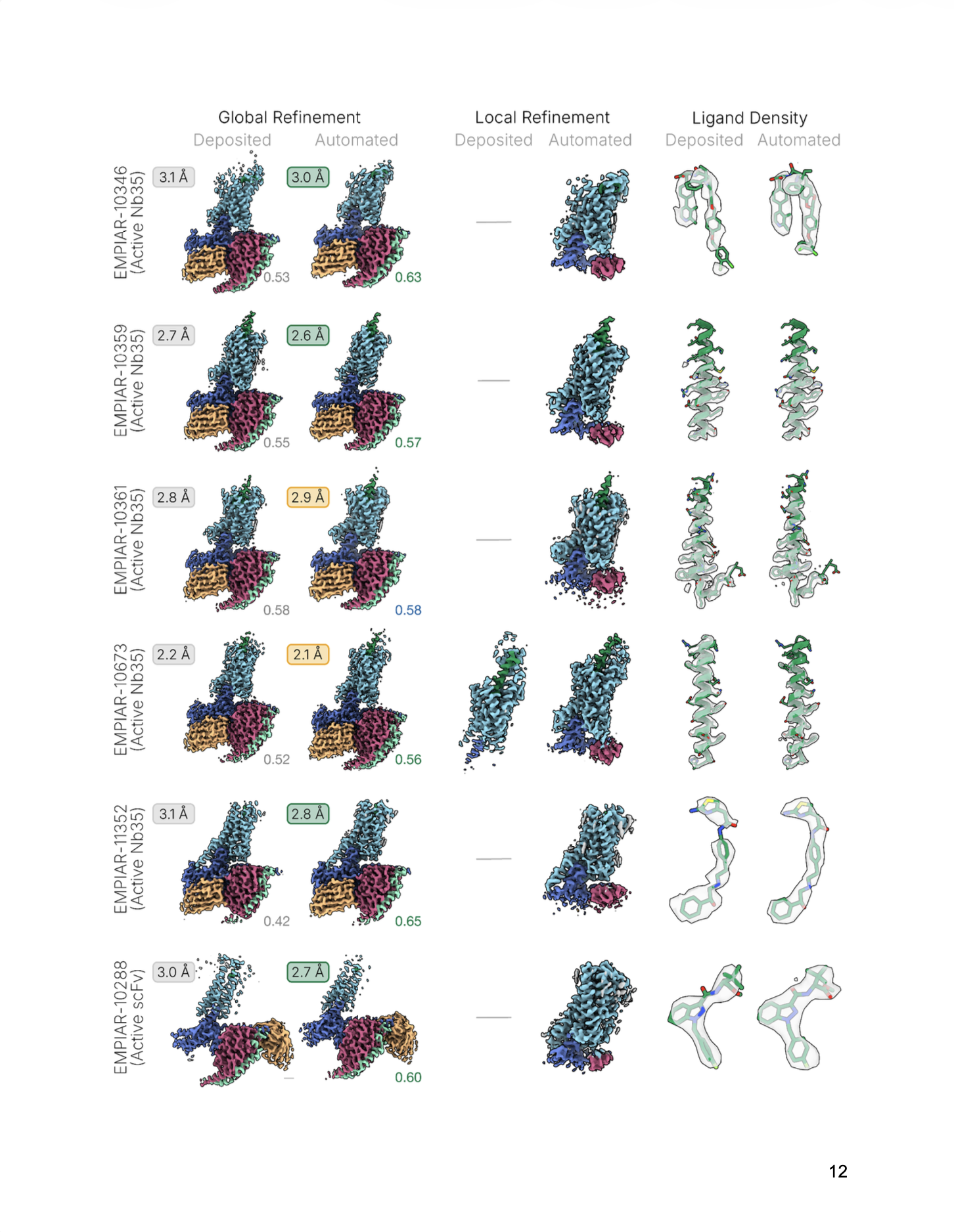
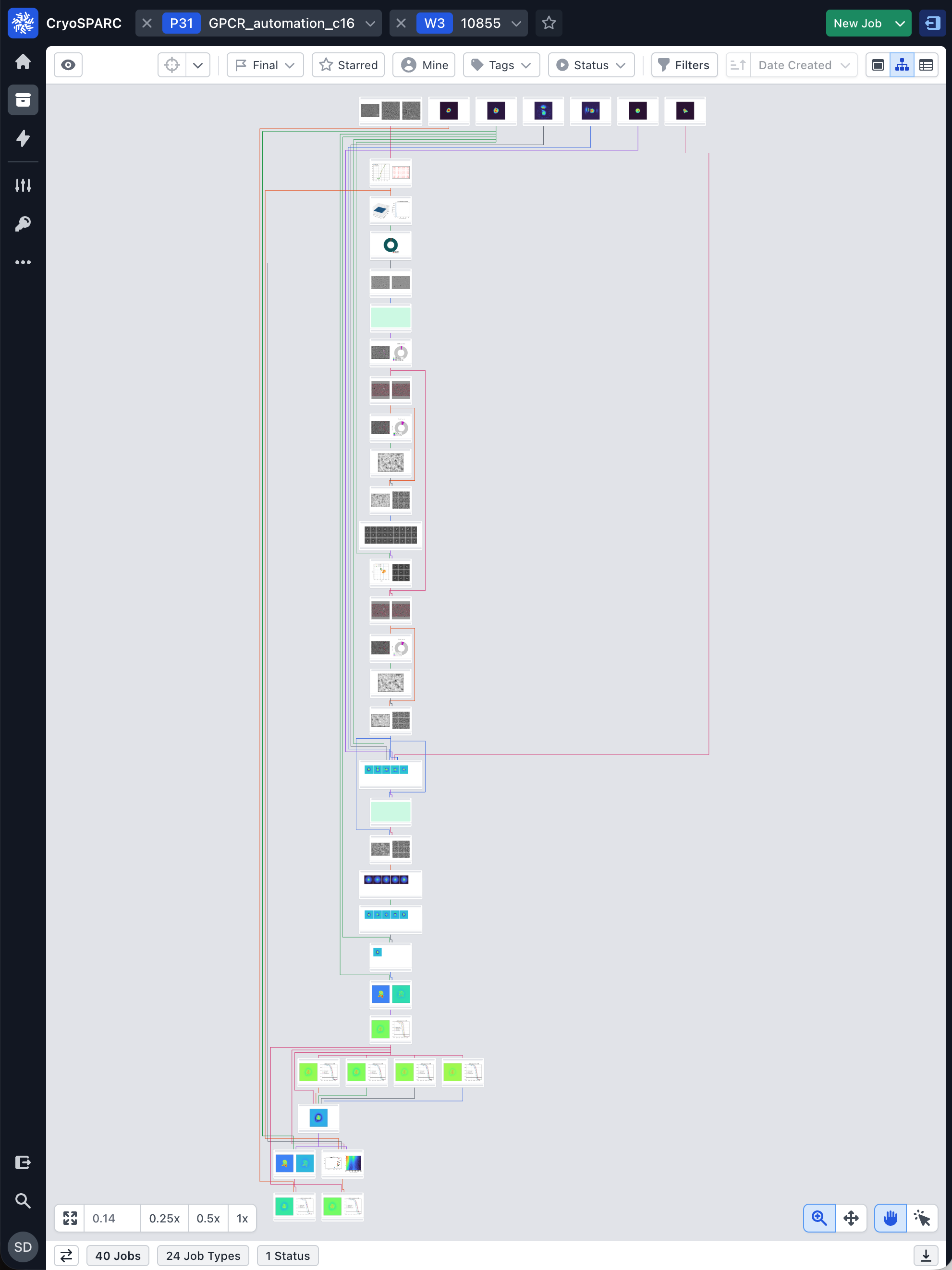
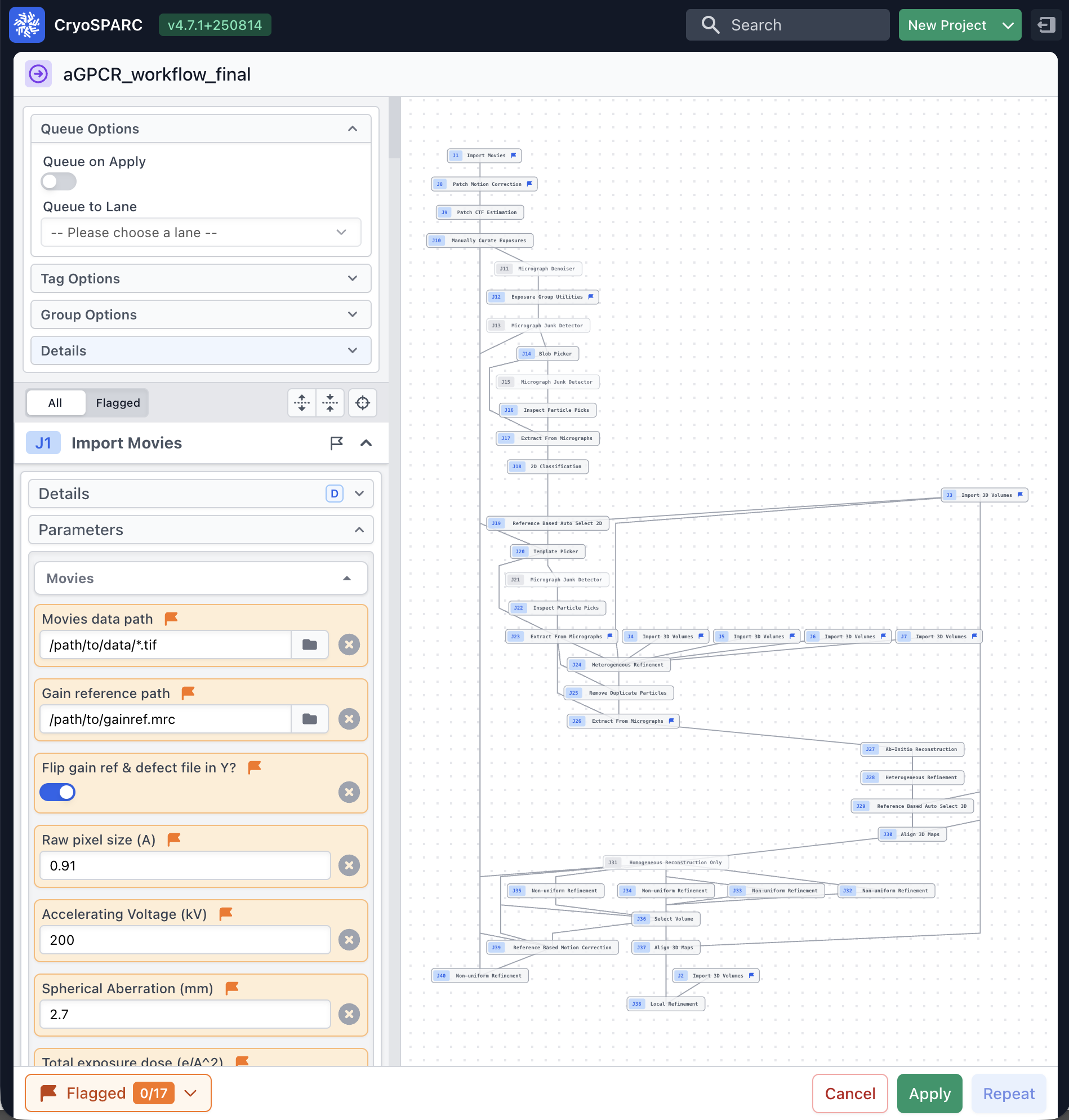
A reusable workflow that encodes best practices—micrograph curation, robust picking, rigorous 3D curation via decoy classification and Ab‑Initio reconstruction, followed by Non‑Uniform and Local Refinement—with optional Reference‑Based Motion Correction.